Meta-Analysis in Neuroimaging
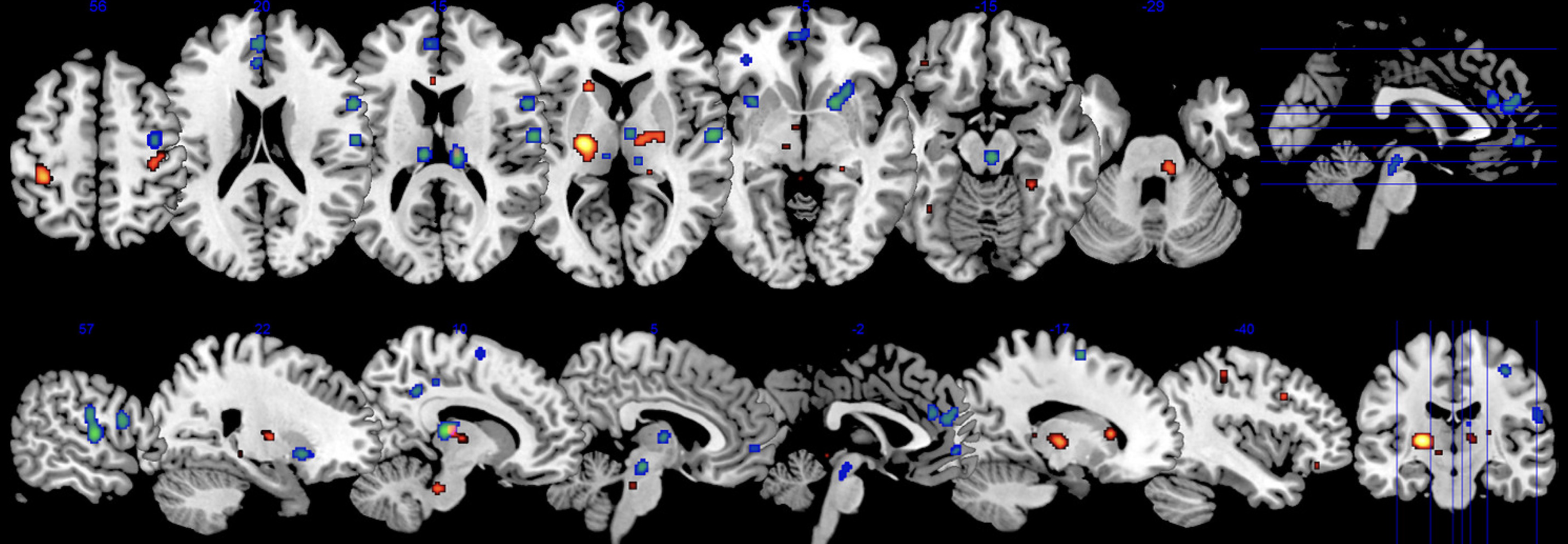
1. Introduction to Meta-Analysis
What is Meta-Analysis?
Meta-analysis is a statistical method that combines results from multiple scientific studies to provide a more comprehensive and reliable conclusion and characterize the degree of agreement across studies. In neuroimaging, it aggregates data from various studies to identify consistent patterns of brain activity.
Importance of Meta-Analysis in Neuroimaging
- Enhanced Statistical Power
- Generalization of Findings
- Identification of Consistent Patterns
Limitations
- Publication Bias (File Drawer Effect): Studies with significant results are more likely to be published, skewing the overall findings.
- Selection Bias: Incorrect inclusion criteria or design flaws in individual studies can affect the validity of the meta-analysis.
2. Types of Meta-Analyses
Image-Based Meta-analysis (IBMA)
“Where do the images converge in space?”
Standards
Uses the full statistic images, and allow the use of hierarchical mixed effects models that account for differing intra-study variance and modeling of random inter-study variation [1].
Limitations
Rarely used because accessing original statistical maps is challenging. Using full image data prevents information loss seen when only specific points (foci) are used. However, until sharing full statistical maps with effect sizes and standard errors becomes common practice, researchers might only have access to lists of specific locations (foci) [1].
IBMA Methods:
- Mixed-Effects General Linear Model (GLM)
- Fixed-Effects General Linear Model (GLM)
- Fisher's IBMA
- Stouffer's IBMA
- z permutation
For more information: IBMA Documentation
Coordinated-Based Meta-analysis (CBMA)
“Where do the coordinates converge in space?”
Standards
Spatial Normalization: Transforming data from its original space to a standardized template (e.g., Talairach or Montreal Neurological Institute (MNI) space). This preprocessing step ensures that coordinates from different studies can be directly compared and combined.
3D Coordinates Reporting: Results are given as x-y-z coordinates, representing the spatial location of peak activations in the brain, following the standardized MNI or Talairach coordinate systems. The x-y-z values correspond to standard spatial directions: left to right (x), back to front (y), and bottom to top (z).
Use of Foci: CBMA uses specific points (foci) as data, making it the most common method in neuroimaging meta-analysis. Foci data are easily accessible in neuroimaging literature, and tools like NeuroSynth, NeuroQuery, and BrainMap facilitate sourcing studies by topic or region of interest.
Main Methods/Approaches
- ACTIVATION LIKELIHOOD ESTIMATION (ALE): creates maps by modeling activation coordinates as 3D Gaussian probability distributions.
- SEED-BASED D-MAPPING (SDM): reconstructs whole-brain statistical maps by combining effect sizes and peak coordinates, resulting in mean maps that are then thresholded for significance.
- MULTILEVEL KERNEL DENSITY ANALYSIS (MKDA): combines density estimates from individual studies and uses a multilevel modeling approach to account for within-study variance and between-study heterogeneity.
Other Methods:
- Kernel Density Analysis (KDA)
- Specific Coactivation Likelihood Estimation (SCALE)
- MKDA Chi2 Extension
3. Conducting a Meta-Analysis
Data Collection
Literature Search: comprehensive search strategies using databases like PubMed, Google Scholar, and neuroimaging-specific repositories.
Inclusion and Exclusion Criteria: defining clear criteria for study selection to ensure consistency and quality.
Data Extraction
Extracting Coordinates: collecting peak activation coordinates from selected studies.
Standardization: converting coordinates to a common brain template space (e.g., MNI or Talairach).
* Alternative option (for ALE meta-analysis): SLEUTH Tool (a Java Runtime Environment 1.8.0 if needed; Java Download). Here for a video tutorial: SLEUTH Tutorial.
Statistical Analysis
Software Tools:
- GingerALE: For ALE meta-analyses. GingerALE Tutorial
- PSI-SDM: For Seed-Based d Mapping meta-analyses.
- NiMARE: Python package for neuroimaging meta-analyses.
- CBMAT GitHub
Visualization of the results
Suggested tools
4. Practical Guide and Tutorials
Tutorial for performing CBMA with GingerALE
Tutorial for performing CBMA with PSI-SDM
5. Resources for Further Learning
Recommended Papers
- Eickhoff, S. B., Bzdok, D., Laird, A. R., Kurth, F., & Fox, P. T. (2012). Activation likelihood estimation meta-analysis revisited. NeuroImage, 59(3), 2349–2361. https://doi.org/10.1016/j.neuroimage.2011.09.017
- Radua, J., Rubia, K., Canales-Rodríguez, E. J., Pomarol-Clotet, E., Fusar-Poli, P., & Mataix-Cols, D. (2014). Anisotropic kernels for coordinate-based meta-analyses of neuroimaging studies. Frontiers in psychiatry, 5, 13. https://doi.org/10.3389/fpsyt.2014.00013
- Albajes-Eizagirre, A., Solanes, A., Fullana, M. A., Ioannidis, J. P. A., Fusar-Poli, P., Torrent, C., Solé, B., Bonnín, C. M., Vieta, E., Mataix-Cols, D., & Radua, J. (2019). Meta-analysis of Voxel-Based Neuroimaging Studies using Seed-based d Mapping with Permutation of Subject Images (SDM-PSI). Journal of visualized experiments : JoVE, (153), 10.3791/59841. https://doi.org/10.3791/59841. https://app.jove.com/t/59841
- Müller, V. I., Cieslik, E. C., Laird, A. R., Fox, P. T., Radua, J., Mataix-Cols, D., Tench, C. R., Yarkoni, T., Nichols, T. E., Turkeltaub, P. E., Wager, T. D., & Eickhoff, S. B. (2018). Ten simple rules for neuroimaging meta-analysis. Neuroscience and Biobehavioral Reviews, 84(April 2017), 151–161. https://doi.org/10.1016/j.neubiorev.2017.11.012
- Manuello, J., Costa, T., Cauda, F., & Liloia, D. (2022). Six actions to improve detection of critical features for neuroimaging coordinate-based meta-analysis preparation. Neuroscience and biobehavioral reviews, 137, 104659. https://doi.org/10.1016/j.neubiorev.2022.104659
- Manuello, J., Liloia, D., Crocetta, A., Cauda, F., & Costa, T. (2024). CBMAT: a MATLAB toolbox for data preparation and post hoc analyses in neuroimaging meta-analyses. Behavior research methods, 56(5), 4325–4335. https://doi.org/10.3758/s13428-023-02185-3
- Samartsidis, P., Montagna, S., Nichols, T. E., & Johnson, T. D. (2017). The coordinate-based meta-analysis of neuroimaging data. Statistical science : a review journal of the Institute of Mathematical Statistics, 32(4), 580–599. https://doi.org/10.1214/17-STS624
Recommended Books and Sites
Online Courses and Lectures
Introductory Lecture Series on Meta-Analysis
Advanced Techniques in Meta-Analysis
References
- Salimi-Khorshidi, G., Smith, S. M., Keltner, J. R., Wager, T. D., & Nichols, T. E. (2009). Meta-analysis of neuroimaging data: A comparison of image-based and coordinate-based pooling of studies. NeuroImage, 45(3), 810–823. https://doi.org/10.1016/j.neuroimage.2008.12.039
- Müller, V. I., Cieslik, E. C., Laird, A. R., Fox, P. T., Radua, J., Mataix-Cols, D., Tench, C. R., Yarkoni, T., Nichols, T. E., Turkeltaub, P. E., Wager, T. D., & Eickhoff, S. B. (2018). Ten simple rules for neuroimaging meta-analysis. Neuroscience and Biobehavioral Reviews, 84, 151–161. https://doi.org/10.1016/j.neubiorev.2017.11.012
- Manuello, J., Costa, T., Cauda, F., & Liloia, D. (2022). Six actions to improve detection of critical features for neuroimaging coordinate-based meta-analysis preparation. https://doi.org/10.1016/j.neubiorev.2022.104659
- Costa, T., Ferraro, M., Manuello, J., Camasio, A., Nani, A., Mancuso, L., Cauda, F., Fox, P. T., & Liloia, D. (2024). Activation Likelihood Estimation Neuroimaging Meta-Analysis: A Powerful Tool for Emotion Research. Psychology Research and Behavior Management, 17, 2331–2345. https://doi.org/10.2147/PRBM.S453035
- Manuello, J., Liloia, D., Crocetta, A., Cauda, F., & Costa, T. (2023). CBMAT: A MATLAB toolbox for data preparation and post hoc analyses in neuroimaging meta-analyses. Behavior Research Methods. https://doi.org/10.3758/s13428-023-02185-3
